I restarted the analyses in early July with the onset of the new Melbourne outbreak in July 2020. The logic behind these charts is that they fill an information gap. Official data sources only give historic data series, and mainstream media typically only give near term predictions based on opinion.
Chart update 13 August 2020
What’s new?
There is a lot new today. I’m have made a change to the model. The Gompertz based model has been overestimating for the past week, and yesterday I stated that its projections into the near future were very likely to be incorrect. This is likely due to transmission dynamics in current stage 4 restrictions being very different to that in the past (for instance, during stage 3 restrictions). My view is that the Gompertz based model, which had done well up to around the introduction of stage 4 restrictions, will simply not be a good description of the case series moving forward. As such, I have “retired” the model. As the aphorism goes, “all models are wrong, but some are useful”.
Moving forward, I am instead cautiously using the Richards’ growth curve, also known as the generalised logistic function, for modelling and projections. This is similar to the Gompertz equation. In fact, the Gompertz equation is a member of the Richards’ growth curves, which is a broad family of curves. I have reasons to believe that it will give more credible projections at this point. This is described in more detail below.
Today’s new number of cases, 292 as per the Australian Government Department of Health on 13/8/2020, is mostly compatible with estimate of the new model.
Projection of new daily cases of COVID-19 with data up to 13 August 2020
What is this?
The image is a chart of the confirmed daily new cases of COVID-19 in Australia, with a projection for the next 2 weeks. The projection is made using a model by fitting the data since 1 June 2020 to a Richards’ growth curve using non-linear regression. The dark green dashed line is the model estimate. The grey dashed lines are the 95% prediction intervals, with the values given at 7 and 14 days into the future. The green gradations can be understood as the degree of uncertainty in the model projections. This model was introduced on 13 August 2020, replacing the previous model that fitted data to a Gompertz equation.
“Richards’ growth curve”? “Gompertz equation”?
Those who have been following these charts would know that the model is constructed by fitting the cumulative daily case numbers to a Gompertz equation. This is an “S-shaped” or sigmoid curve. There were two rationales in doing this. The first is that the Gompertz function does describe well certain types of biological growth, including epidemics. The equation itself is the solution to a specific differential equation that has some plausibility with regards to case growth in an epidemic. The second more empirical rationale is that Chinese authors found that the Gompertz equation appeared to describe the growth of COVID-19 cases in parts of China well (Jia et al. arXiv:2003.05447v2 [q-bio.PE]). I personally found this was the case in the March-April outbreaks in Australian and New Zealand.
However, the Gompertz equation does have limitation. For one, the inflexion point of the curve is fixed. The inflexion point, is where we would see the peak number of new cases a day. This is well demonstrated in the NZ outbreak of March 2020 (see below).
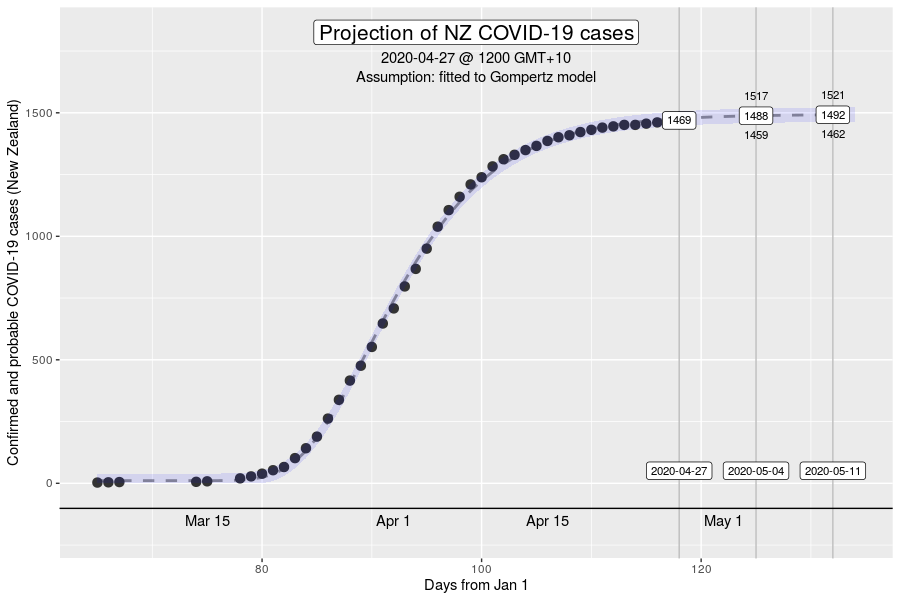
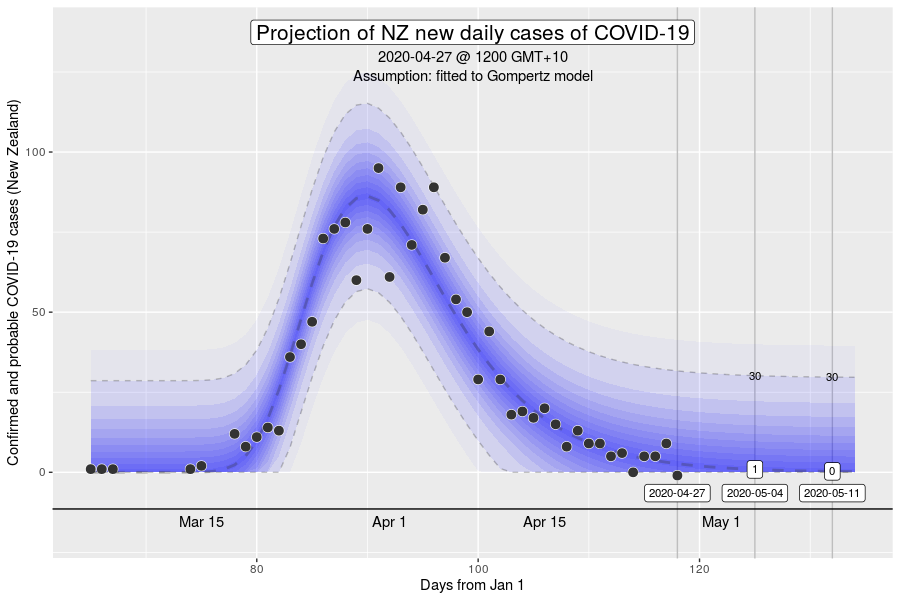
The inflexion point of the Gompertz function at 37% of the final total number of cases in an outbreak (exactly at the ratio of 1/e, where e is Euler’s number). While this is what we expect might in an outbreak where the growth dynamics remains relatively constant, it will be a poor description if the intensity of interventions to control transmission increases over time (e.g., with the stage 4 restrictions).
The Richards’ growth curve (or the generalised logistic function) is a broad family of sigmoid curves, and can describe curves with inflexion points that vary. It too has been demonstrated to have utility in modelling COVID-19 outbreaks (Lee et al. PLoS One 2020 doi: 10.1371/journal.pone.0236860). A consequence of this freedom, however, is that IF data is well describe by the Gompertz equation, there will be greater uncertainty and statistical error using a Richards’ curve. This is not just a theoretical concern. When I use the Richards’ growth curve to model data from earlier in the Melbourne outbreak, it tends to result in very wide prediction intervals in near future projections, with an estimate that biases towards under-estimates. Effectively, projections from a Richards’ growth curve using early data result in overly optimistic estimates of how soon the inflexion point or the “peak” will come.
So why now? Firstly, we have crossed the peak/passed the inflexion point already. My intuition is that the more flexible nature of the Richards’ curve now that data exists for a larger range of the curve, will lead to much better performance than using a similar method with the Gompertz equation. Moreover, it is pretty clear that the projections from the Gompertz equation are not credible in the near future, due to it’s fixed inflexion point, and that the actual case curve has been “bent down” through interventions.
Comparison between the Gompertz and Richards’ growth curve model projections, along with smoothed data trends (moving average vs GAM) with data up to 13 August 2020
Where the Gompertz model is almost certainly overestimating currently, Richards’ growth curve model might be underestimating. These are important biases to keep in mind.
How have the model projections changed over the month?
The video demonstrates how the projections have evolved over time as new daily data have become available. This can give a better sense of where we are headed, given that the model cannot account for changes in context (e.g., policy changes, changes in testing rates, etc.) Please note the model change that occurred on 13 August 2020 (Gompertz = blue, Richards’ = green).
My interpretation
There is a clear and substantial downward change in trajectory in the past few days. This is great. This is highly likely to be the result of the stage 4 restrictions. To visually see how stage 3 and then stage 4 restrictions affected the long term trajectory of the outbreak, we can use the “asymptote” of the Gompertz model, which is one of its parameters. The interpretation of this value is that it is the projected final size of the outbreak in terms of the total number of cases. Note: the value of this parameter must be seen as highly speculative and be understood as conceptual “ballpark” terms.
Each data point is the projected final size of the outbreak based on the model with data available up to that day. The trend line is from a generalised additive model, with 95% confidence intervals of the trend.
As can be seen prior to stage 3 restrictions, new daily data could be understood as showing something quite alarming. As the model was updated with each day’s new number of COVID-19 cases, the final potential size of the outbreak was getting larger and larger. By mid-June, the model suggests up to 60 to 90 thousand people might eventually become infected. At around a week after stage 3 restrictions were implemented, this completely around, but then plateaus. It would seem that if we stayed only at stage 3 restriction, the potential final size of the outbreak will still be in the order of 40 to 50 thousand people. Since stage 4 restrictions, it’s clear that projected final size of the outbreak is starting to fall again, indicating that transmission is being suppressed further.
Despite some negative voices in the free media, it is my view that it is categorically clear that both stage 3 and stage 4 restrictions had substantial and important effects in curtailing the outbreak. We must all play our part to try to limit transmission where we can. We are all in this together.
Want to know more?
Primary data source is the Australian Government Department of Health COVID-19 website for daily new cases. Analysis done using RStudio Cloud using R version 4.0.2.
Today’s charts
Data: au_covid
R code: richards_model
